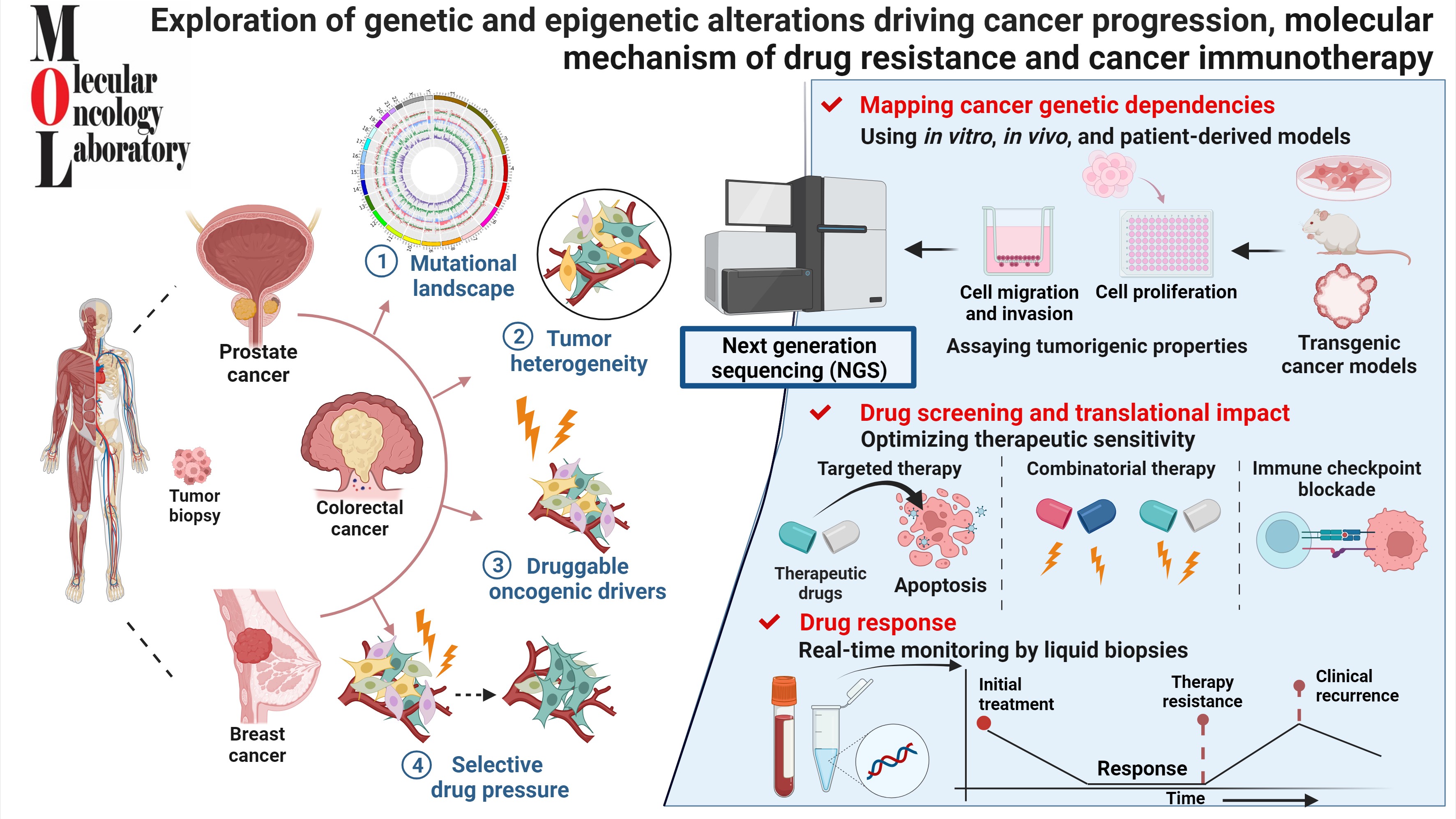
Research Description
My group is interested in exploring the genetic/epigenetic changes that initiate cancer and its progression by employing novel strategies and approaches. We also aim to investigate the molecular events that drive cancer progression and resistance towards chemotherapeutic drugs, in hopes that these discoveries can lead to the development of more effective therapies that target the specific causative pathways/alterations.
Prostate Cancer
Molecular characterization of Indian Prostate Cancer
Our research group in collaboration with GSVM medical college, Kanpur; King George’s Medical University, Lucknow and All India Institute of Medical Sciences, New Delhi for the first time screened Indian prostate cancer (PCa) cohort for the known genetic alterations. Our group for the first time characterized Indian PCa patients’ specimens for ERG, ETV1, ETV4 and RAF kinase genetic rearrangements, SPINK1 over-expression and PTEN deletion by employing immunohistochemistry (IHC) and Fluorescence in-situ Hybridization (FISH). We revealed high recurrence of TMPRSS2-ERG (~50% recurrent) and SPINK1 over-expression (~12% recurrent) among Indian PCa patients. Most importantly, we found higher incidence of RAF genetic rearrangements among Indian prostate cancer patients (~6% in Indian PCa as compared to ~2% in Caucasian), indicating that these patients may respond to the FDA-approved RAF kinase inhibitors or MEK inhibitors which are already available in the clinic (Prostate, 2015). This work represents the first comprehensive molecular subtyping of prostate cancer cohort from the Indian subcontinent, which has a huge potential for direct diagnostic, tailored therapeutic intervention and translational impact.
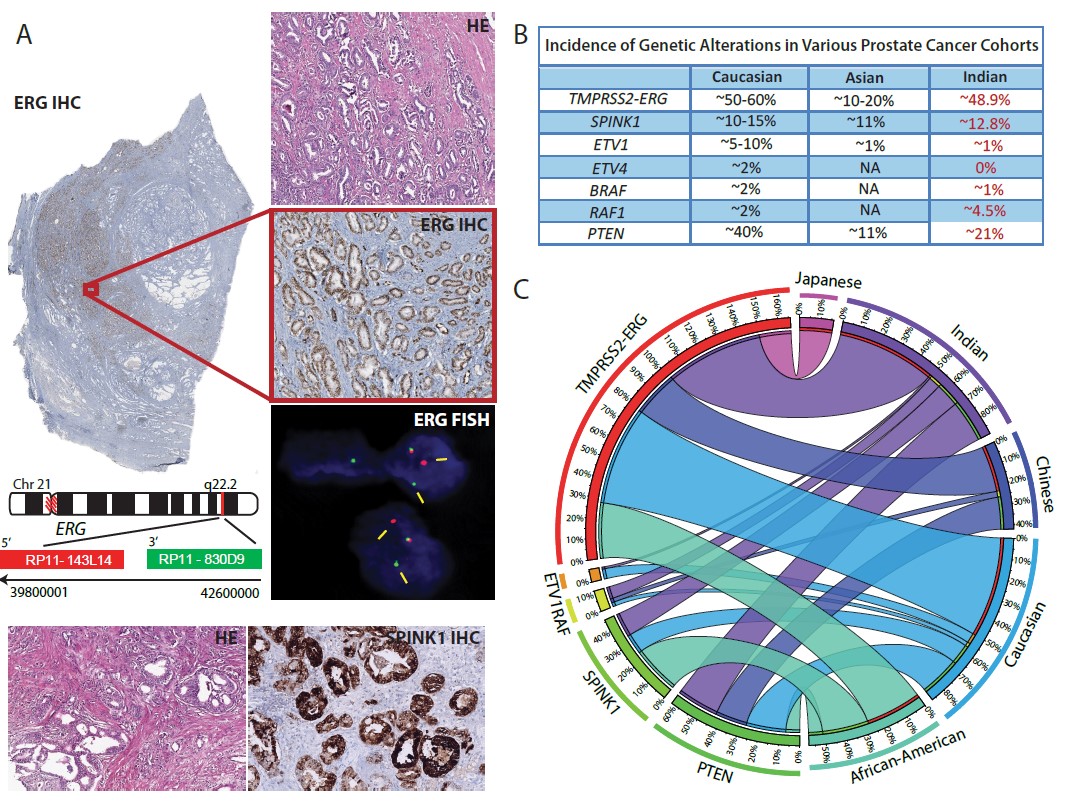
(A) IHC staining for ERG rearrangement and SPINK1 in Indian PCa patients. (B) Summary of molecular subtypes in prostate cancer. (C) Circos depicting racial disparity associated with molecular subtypes of prostate cancer (Trends in Cancer, 2016).
Exploration of Novel Molecular Drivers and Mechanistic Circuitry of Indian Prostate Cancer
Prostate cancer has been known to evolve from diverse genetic alterations including mutations, gene fusions/rearrangements, amplifications/deletions, and other aberrations that perturb gene expression levels. The major drift in field of prostate cancer genomics has been the sub-classification of tumors into distinct molecular subtypes. Emerging high throughput technologies such as Next Generation Sequencing will dictate the developments in personalized genomic medicine in the near future. Although majority of Our group’s major focus of research is to fully understand the entire spectrum of genetic/epigenetic aberrations that occur in cancer patients specifically in the Indian subcontinent. Our previous efforts successfully delineated the common molecular subtypes for Indian PCa patients using a targeted IHC/FISH-based approaches. We to explore the comprehensive mutational landscape of these patients representing the entire disease-spectrum (primary to metastatic disease), understand the pathobiology of the newly identified mutation(s) and identify actionable alterations.
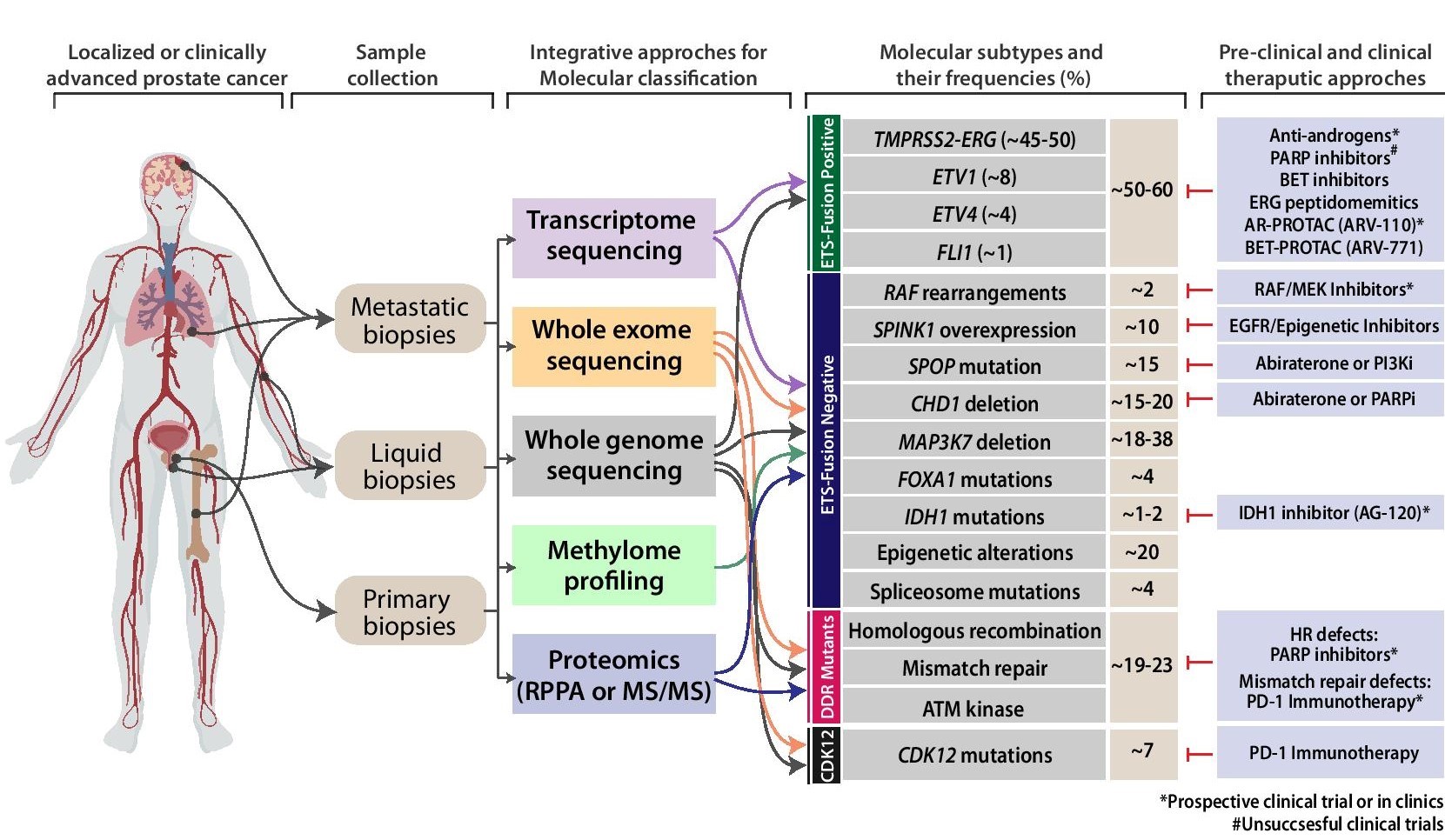
Considering racial disparities and clinical-treatment options available in India, exploring mutational-profiles of patient representing prostate cancer disease progression is critical for understanding the pathobiology and in redefining therapeutic targets for these patients. Major focus of our current research includes:
- Genome wide screen of frequently mutated and hotspot regions associated with prostate cancer risk in Indian patient samples.
- Integrative Sequencing for comprehensive identification of clinically significant alterations in Indian Prostate Cancer.
- Development of prostate cancer patient-derived organoids from biopsies and utilize them genomic characterization and potential drug screen.
- Characterization of exosome-encapsulated coding and non-coding RNAs secreted during prostate cancer using liquid biopsies.
Investigating the role of Homeobox genes in prostate cancer progression
Homeobox genes are the master regulators encoding transcription factors (TFs) which play a crucial role in the embryonic development. These TFs contain a conserved DNA binding domain known as homeodomain. Dysregulation of homeobox gene family including HOX and NKX3.1 have been reported in PCa progression. Distal-less homeobox (DLX) gene family members have been identified as early diagnostic biomarkers in PCa patients, whose oncogenic potential still remains elusive. We intend to unravel the molecular mechanism behind DLX gene family mediated PCa oncogenicity, which will open up avenues for novel therapeutic interventions targeting this molecular subtype of PCa.