~Research~
-
Origin of the Nonlinear Structural and Mechanical Properties in Oppositely Curved Lipid Mixtures
Structural and mechanical properties of membranes such as thickness, tail order, bending modulus and curvature energetics play crucial role in controlling various cellular functions that depend on the local lipid organization and membrane reshaping. While behavior of these biophysical properties are well understood in single component membranes, very little is known about how do they change in the mixed lipid membranes. Often various properties of the mixed lipid bilayers are assumed to change linearly with the mole fractions of the constituent lipids which, however, is true for “ideal” mixing only. In this study, using molecular dynamics simulations, we show that structural and mechanical properties of binary lipid mixture change nonlinearly with the lipid mole fractions, and the strength of the nonlinearity depends on two factors - spontaneous curvature difference and locally inhomogeneous interactions between the lipid components.
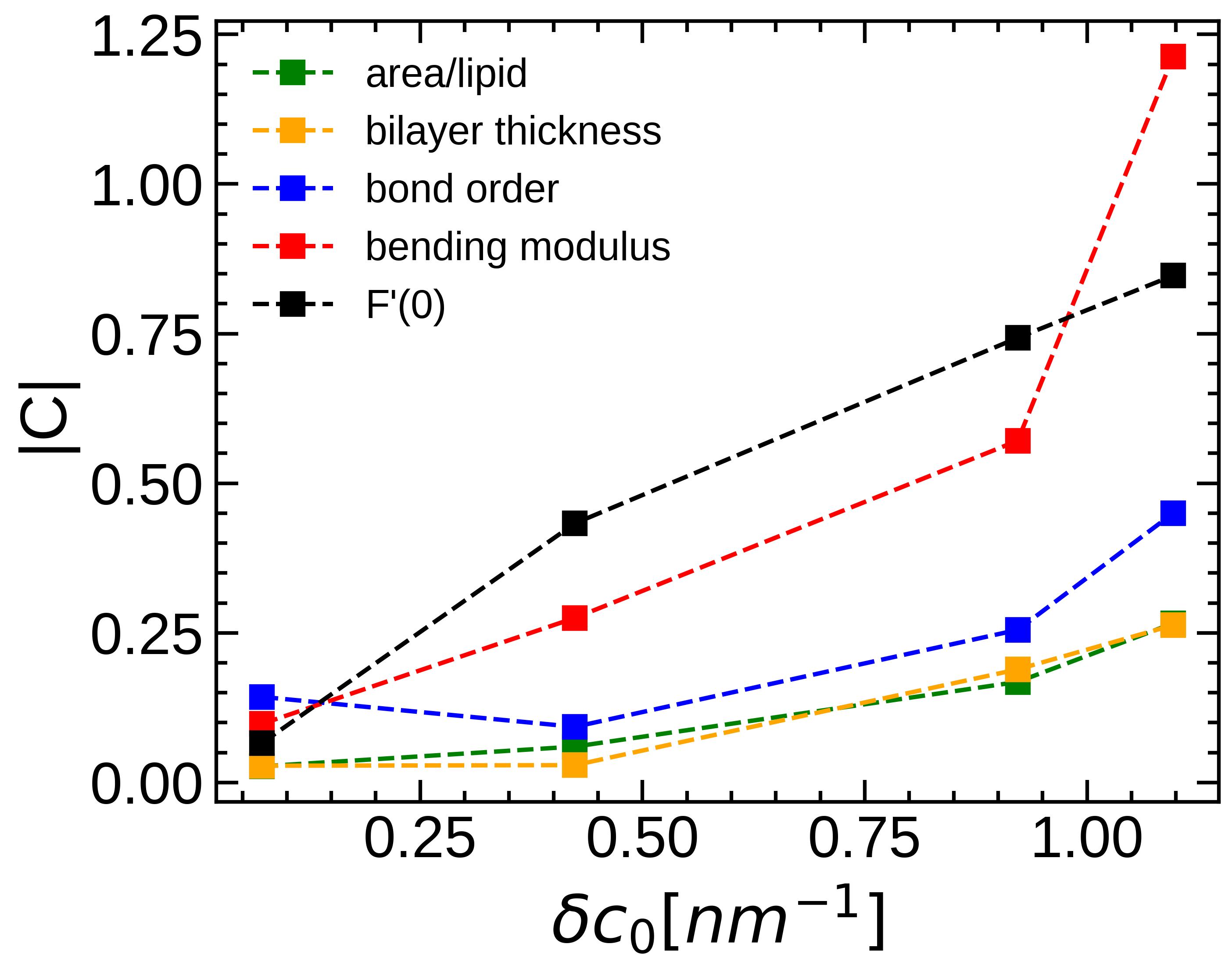
[1] Shivam Gupta, Jatin Soni, Awneesh Kumar and Taraknath Mandal. "Origin of the nonlinear structural and mechanical properties in oppositely curved lipid mixtures." J. Chem. Phys.(2023).
Membrane bending by protein crowding: A case study with the epsin N-terminal homology domain
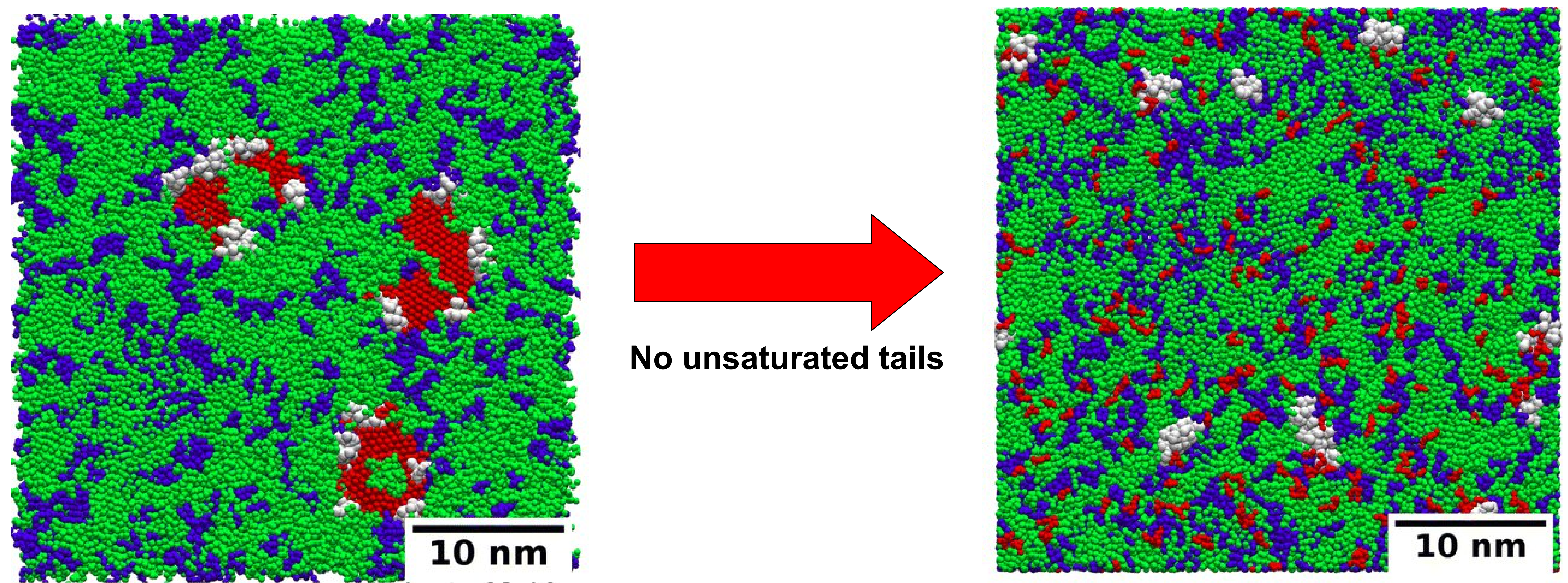
Membrane bending or remodeling is essential for numerous cellular processes including exocytosis, endocytosis, cell division, wound repair, organelle biogenesis, cytokinesis, membrane fusion and fission among many others. In many of these processes, cell membranes adopt highly curved shapes. The mechanisms by which peripheral membrane proteins generate curvature is currently an active area of research. One of the proposed mechanisms is amphipathic insertion or the ‘wedge’ mechanism in which the protein shallowly inserts an amphipathic helix inside the membrane to drive the curvature. However, recent experimental studies have challenged the efficiency of the ‘wedge’ mechanism as it requires unusual protein densities. These studies proposed an alternative mechanism, namely ‘protein-crowding’, in which the lateral pressure generated by the random collisions among the membrane bound proteins drives the bending. We employ atomistic and coarse-grained molecular dynamics simulations to investigate the effects of amphipathic insertion and protein crowding on the membrane surface. Considering epsin N-terminal homology domain as a model protein, we show that amphipathic insertion is not essential for membrane bending. Our results suggest that ENTH domains can aggregate on the membrane surface by employing another structured region. And this protein crowding decreases the cohesive energy of the lipid tails which causes a significant decrease in the membrane bending rigidity. The ENTH domain can generate a similar degree of membrane curvature irrespective of the activity of its H0 helix.

[1] Taraknath Mandal, Shivam Gupta and Jatin Soni. "Simulation study of membrane bending by protein crowding: a case study with the epsin N-terminal homology domain." Soft Matter (2023).
Functional membrane microdomains (FMMs) formation in bacterial membranes
Recent experimental studies revealed that functional membrane microdomains (FMMs) are formed in prokaryotic cells which are structurally and functionally similar to the lipid rafts formed in eukaryotic cells. We employ coarse-grained molecular dynamics simulations to investigate the mechanism of domain formation and its physiochemical properties in a model methicillin-resistant staphylococcus aureus (MRSA) cell membrane. We find that domains are formed through lateral segregation of staphyloxanthin (STX), a carotenoid which shields the bacteria from the host's immune because of its antioxidant nature. Simulation results suggest that membrane integrity increases with the size of the domain and various membrane domain proteins such as flotillin-like protein floA and penicillin binding protein (PBP2a) preferentially bind with the STX and accumulate in the membrane domain which is consistent with the recent experimental results.
[1] Shivam Gupta and Taraknath Mandal. "Simulation study of domain formation in a model bacterial membrane." Physical Chemistry Chemical Physics (2022).